Virtual Chunks
Arraylake's native data model is based on Zarr Version 3. However, through Icechunk's support for Virtual Chunks, Arraylake can ingest a wide range of other array formats, including:
- "Native" Zarr Version 2/3
- HDF5
- NetCDF3
- NetCDF4
- GRIB
- TIFF / GeoTIFF / COG (Cloud-Optimized GeoTIFF)
Importantly, these files do not have to be copied in order to be used in Arraylake. If you have existing data in these formats already in the cloud, Arraylake can store an index pointing to the chunks in those files. These are called virtual chunks. They look and feel like regular Icechunk chunks, but the chunk data lives in the original file, rather than inside your Icechunk Repository.
Bucket Configuration
The original files do not have to live in the same bucket as your Icechunk Repository,
but they must live in a location for which your org has a BucketConfig set up (see Managing Storage).
Note that you can create BucketConfigs for buckets which you do not have write access to, such as those owned by external organisations.
This means you can ingest read-only references to data provided by other organisations, such as the public sector.
Simply create a BucketConfig the same way you would for non-virtual data.
Currently only anonymous access buckets are supported for use with virtual chunks in arraylake.
If you create an Arraylake repo containing virtual chunks referencing files in a bucket with any other type of authentication method (e.g. HMAC or delegated credentials), an error will be raised when you try to authorize access to the virtual chunk data. This is for security reasons.
Using virtual chunks with Icechunk requires a more recent version of Icechunk: >=v1.1.16.
Virtual Chunk Containers
Virtual Chunk Containers tell Icechunk how to access storage locations containing virtual chunk data.
Icechunk needs to know the bucket prefixes containing the data, which will be of a form like "s3://my-bucket/someprefix/".
A user with write access to the Arraylake repo must set these prefixes.
Icechunk supports some types of virtual chunk container prefixes which are forbidden in Arraylake.
The forbidden prefixes are file:///, memory://, http://, and https://.
If you create an Arraylake repo with a virtual chunk container that uses a forbidden prefix, an error will be raised when you try to authorize access to the virtual chunk data. Again this is for security reasons.
Authorizing Virtual Chunk Access
For security reasons,
Icechunk requires you to explicitly authorize access to virtual chunks, by passing credentials explicitly.
Arraylake currently also requires you to explicitly authorize access at write time, by passing the authorize_virtual_chunk_access kwarg to client.create_repo/client.get_repo.
The difference is that while Icechunk's authorization requires you to generate and pass bucket credentials yourself, Arraylake's authorization only requires you to authorize access to a particular bucket config, and Arraylake handles generating the specific credentials for you.
To authorize reading virtual chunks from an Arraylake repo, two things need to happen:
- A user with write access to the Arraylake repo must specify the authorized virtual chunk prefixes up front.
This can be done either at repo creation time (by passing
authorize_virtual_chunk_accesstoclient.create_repo()) or later on (by callingclient.authorize_virtual_chunk_access()). - The user wishing to read the virtual chunk data back must also authorize access, this time by passing the
authorize_virtual_chunk_accesskwarg toclient.get_repo(). But by default arraylake will do this for you - "auto-discovering" the correct kwarg.
The authorize_virtual_chunk_access kwarg maps the virtual chunk container prefix expressing the location of the virtual chunk data (e.g. s3://my-bucket/legacy/) to the bucket config you set up for that storage location.
For example authorize_virtual_chunk_access={"s3://my-bucket/legacy/": "netcdf-data"},
where "netcdf-data" is the nickname of a BucketConfig for a bucket containing some netCDF files, which are all inside the legacy/ bucket prefix.
Viewing Virtual Chunk Containers
Once set, you can view the authorized virtual chunk containers either through the web app or by using the python client.
- Web App
- Python
- Python (asyncio)
In the web app, navigate to the repository's Settings > General page. If the repository has virtual chunk containers configured, you will see a Virtual Chunk Buckets section below the Repo Bucket section.
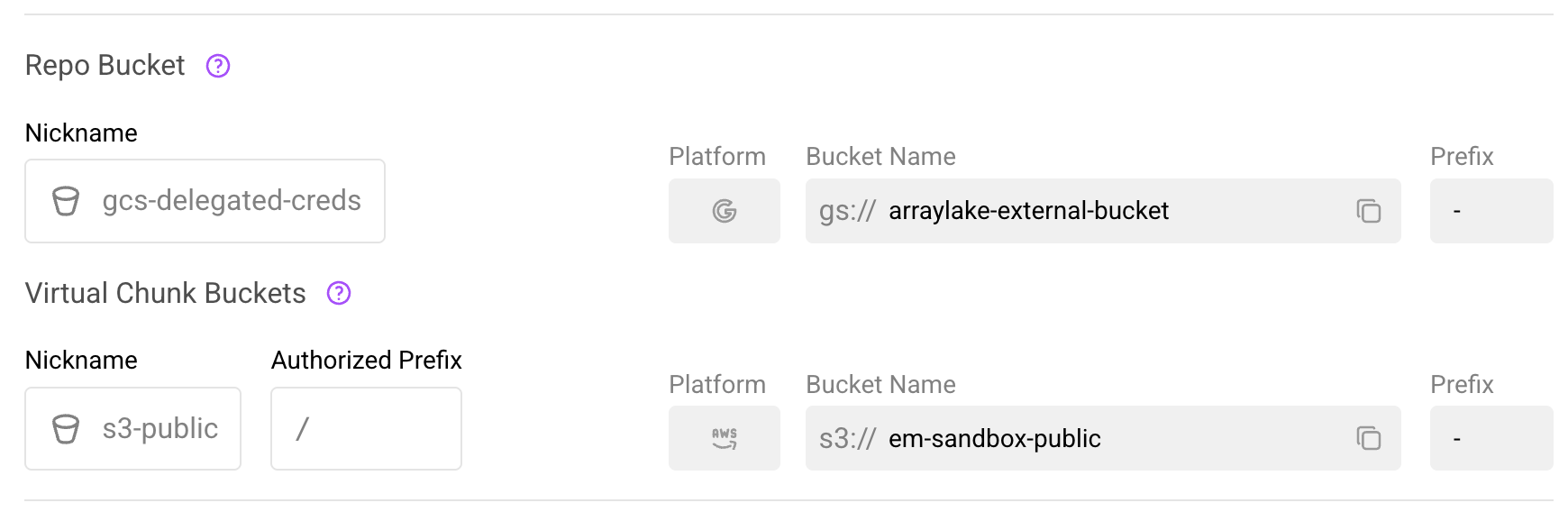
The Virtual Chunk Buckets section in repository settings.
This section displays:
- Nickname: The bucket configuration nickname
- Authorized Prefix: The path within the bucket that is authorized for virtual chunk access
- Platform, Bucket Name, Prefix: Details of the underlying bucket configuration
from arraylake import Client
client = Client()
# Get virtual chunk containers for a repo
vcc_mapping = client.get_virtual_chunk_containers("earthmover/my-repo")
print(vcc_mapping)
{'s3://noaa-gefs-retrospective/GEFSv12/reforecast/': 'noaa-gefs'}
from arraylake import AsyncClient
aclient = AsyncClient()
# Get virtual chunk containers for a repo
vcc_mapping = await aclient.get_virtual_chunk_containers("earthmover/my-repo")
print(vcc_mapping)
{'s3://noaa-gefs-retrospective/GEFSv12/reforecast/': 'noaa-gefs'}
Updating Virtual Chunk Containers
Users with write access to a repository can add or remove virtual chunk containers at any time.
- Web App
- Python
- Python (asyncio)
- Navigate to the repository's Settings > General page
- Click the Edit button (pencil icon) next to the "Virtual Chunk Buckets" heading
- To add a new virtual chunk bucket:
- Click Add Virtual Chunk Bucket
- Select a bucket from the dropdown (only buckets with anonymous access are available)
- Optionally specify an additional prefix path within the bucket
- The new entry will appear with a dashed border indicating it's pending
- To remove a virtual chunk bucket, click the trash icon next to the entry and confirm the removal
- Click Save Changes to apply your modifications
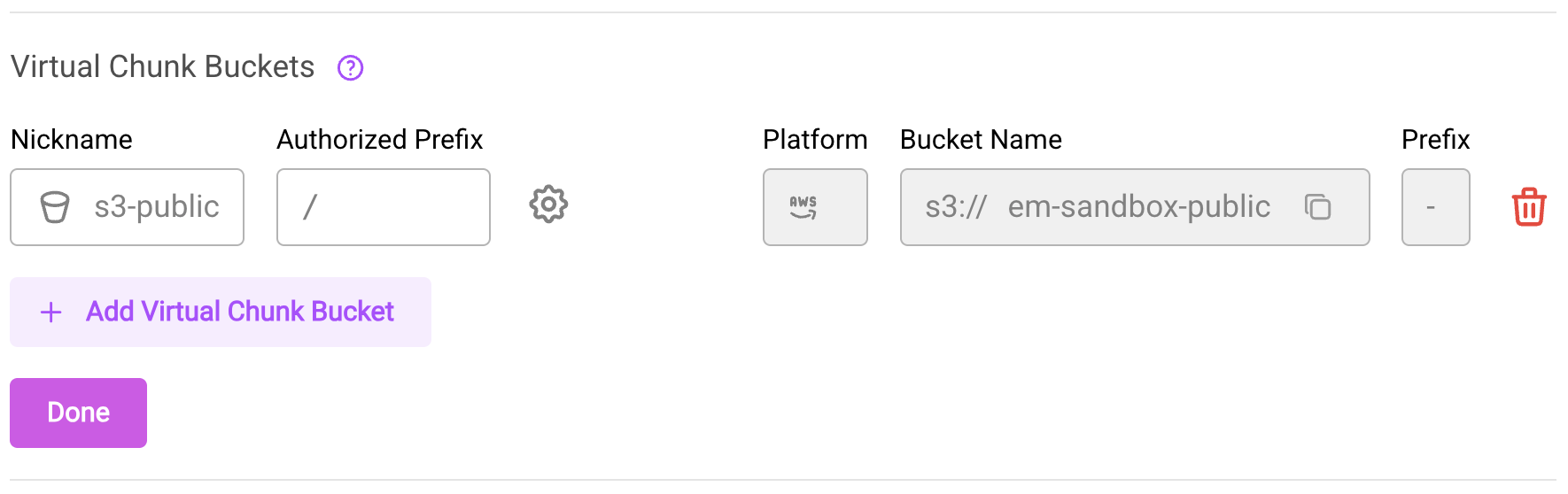
Editing the Virtual Chunk Buckets section in repository settings.
Only bucket configurations with anonymous access can be used for virtual chunks. If you don't see the bucket you need in the dropdown, check that it has been configured with anonymous access in your organization's bucket settings.
You can pass authorize_virtual_chunk_access to create_repo to set the virtual chunk containers immediately:
from arraylake import Client
client = Client()
client.create_repo(
"earthmover/my-repo",
authorize_virtual_chunk_access={"s3://another-bucket/data/": "another-bucket-config"},
)
Or use set_virtual_chunk_containers to reset the virtual chunk containers at any point later on:
from arraylake import Client
client = Client()
client.set_virtual_chunk_containers(
"earthmover/my-repo",
authorize_virtual_chunk_access={"s3://another-bucket/data/": "another-bucket-config"},
)
You can pass authorize_virtual_chunk_access to create_repo to set the virtual chunk containers immediately:
from arraylake import AsyncClient
aclient = AsyncClient()
await aclient.create_repo(
"earthmover/my-repo",
authorize_virtual_chunk_access={"s3://another-bucket/data/": "another-bucket-config"},
)
Or use set_virtual_chunk_containers to reset the virtual chunk containers at any point later on:
from arraylake import AsyncClient
aclient = AsyncClient()
await aclient.set_virtual_chunk_containers(
"earthmover/my-repo",
authorize_virtual_chunk_access={"s3://another-bucket/data/": "another-bucket-config"},
)
Ingesting Virtual Data
Let's walk through an example of ingesting some data into Arraylake as virtual chunks. We will create a mirror of the NOAA GEFS Re-forecast data available on the AWS Open Data Registry.
This involves ingesting some netCDF4 data in a public anonymous access bucket, and using the VirtualiZarr package to parse the contents of the files.
Create Bucket Config
First we need create a bucket config for the location of the data we wish to ingest. We can do this via the web app or via the python client, but here we'll use the python client:
from arraylake import Client
client = Client()
client.create_bucket_config(
org="earthmover",
nickname="noaa-gefs",
uri="s3://noaa-gefs-retrospective/",
prefix="GEFSv12/reforecast"
extra_config={'region_name': 'us-east-1'}
)
Create Repo
Now we need to create an Arraylake repo, and authorize the virtual chunk container.
ic_repo = client.create_repo(
"earthmover/gefs-mirror",
description="NOAA data!",
metadata={"type": ["weather"]},
authorize_virtual_chunk_access={
"s3://noaa-gefs-retrospective/GEFSv12/reforecast/": "noaa-gefs",
}
)
(Recall that "s3://noaa-gefs-retrospective/GEFSv12/reforecast/" is the bucket prefix to which we want to authorize access,
and noaa-gefs is the nickname of the bucket config we set up for that bucket.)
Create virtual references and commit them to Icechunk
We use the VirtualiZarr package to parse the netCDF file and commit the virtual references to Icechunk.
session = ic_repo.writable_session("main")
vds = vz.open_virtual_dataset(path, parser=vz.HDFParser())
vds.vz.to_icechunk(session.store)
session.commit("wrote some virtual references!")
See data in the web app
We should now be able to see the data that we ingested in the Arraylake web app.
Read the data back
Finally, we can now access the data we ingested!
Notice users do not need to authorize at read-time by passing authorize_virtual_chunk_access, though if they want to override by passing it they can.
import xarray as xr
ic_repo = client.get_repo(
"earthmover/gefs-mirror",
)
session = ic_repo.readonly_session("main")
ds = xr.open_zarr(session.store)
ds["some_variable"].plot()
Advanced ingestion
More complicated virtual data ingestion tasks are possible, including:
- Concatenating references to multiple files before ingestion,
- Appending to existing datasets along a specific dimension (e.g.
time), - Ingesting other data formats.
For information about such workflows, please see the Icechunk documentation on Virtual Datasets and the VirtualiZarr documentation.