Use Arraylake with Xarray
In Manage Zarr Data, we saw how to interact with Arraylake using the Zarr python API.
On this page, we will see how to read and write Xarray Datasets to / from Arraylake.
import xarray as xr
from arraylake import Client
In the previous example, we learned how to create repos.
Here we'll use a new repo called earthmover/xarray-tutorial-datasets.
client = Client()
repo = client.create_repo("earthmover/xarray-tutorial-datasets")
repo.status()
🧊 Using repo earthmover/xarray-tutorial-datasets
📌 Base commit is None
📟 Session bbd3b89b57a545638fc1fa825795f772 started at 2024-11-25T18:57:32.424000
🌿 On branch main
No changes in current session
Xarray comes with a number of tutorial datasets. Let's store one of these to Arraylake.
First we'll create a group to hold these datasets. (This is optional. We could also just put them in the root group.)
group = repo.root_group.create_group("xarray_tutorial_datasets")
Load an Xarray Dataset
Now let's load one of the Xarray tutorial datasets.
ds = xr.tutorial.open_dataset("air_temperature")
print(ds)
<xarray.Dataset> Size: 31MB
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float64 31MB ...
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...
Many NetCDF datasets come with variable "encoding" which specifies details of how to store the data on disk. We generally want to remove these before saving data to Zarr and let Xarray choose the encoding.
ds = ds.drop_encoding()
Store to Arraylake
Now let's store it in Arraylake. Use the following syntax.
ds.to_zarr(
repo.store,
group="xarray_tutorial_datasets/air_temperature",
zarr_version=3,
mode="w"
)
<xarray.backends.zarr.ZarrStore at 0x1562f42c0>
Let's make a commit to save this update.
cid = repo.commit("Wrote Xarray dataset to Arraylake")
cid
6744c97898c20ad7df9b3666
Open our Data using Zarr
Although our dataset was written from Xarray, it is also a valid standalone Zarr dataset. It can be helpful to open your data directly using Zarr in order to understand what has been stored. Compared to Xarray, Zarr offers a more "low-level" view on the raw data, without any coordinates or computational methods.
The following command shows the hierarchy of the entire repo.
repo.tree().__rich__()
/
└── 📁 xarray_tutorial_datasets
└── 📁 air_temperature
├── 🇦 air (2920, 25, 53) float32
├── 🇦 lat (25,) float32
├── 🇦 lon (53,) float32
└── 🇦 time (2920,) int64
In order to see more details about chunks, compression, etc. we can use .info.
repo.root_group.xarray_tutorial_datasets.air_temperature.air.info
| Name | /xarray_tutorial_datasets/air_temperature/air |
|---|---|
| Type | zarr.core.Array |
| Data type | float64 |
| Shape | (2920, 25, 53) |
| Chunk shape | (730, 7, 14) |
| Order | C |
| Read-only | False |
| Compressor | Blosc(cname='lz4', clevel=5, shuffle=SHUFFLE, blocksize=0) |
| Store type | arraylake.repo.ArraylakeStore |
| No. bytes | 30952000 (29.5M) |
| No. bytes stored | 19031132 (18.1M) |
| Storage ratio | 1.6 |
| Chunks initialized | 64/64 |
Because we did not specify chunks explicitly when we stored the data, Zarr decided for us. It picked the shape (730, 7, 27) for this array. This may or may not be optimal, depending on our access patterns.
Customize Chunk Size
Let's rewrite the array using new chunks which we specify. (See Xarray Docs for more details on how to specify chunks when writing Zarr data.) The chunks we chose are contiguous in the spatial dimensions.
encoding = {"air": {"chunks": (100, 25, 53)}}
ds.to_zarr(
repo.store,
group="xarray_tutorial_datasets/air_temperature",
zarr_version=3,
mode="w",
encoding=encoding
)
<xarray.backends.zarr.ZarrStore at 0x155f06ec0>
We'll commit the changes to create a new version of the array.
repo.commit("Changed chunking on air temperature dataset")
6744c9f10e86f720cdcd34d0
repo.root_group.xarray_tutorial_datasets.air_temperature.air.chunks
(100, 25, 53)
Note that we can roll back to the earlier version and see the original chunks.
repo.checkout(cid)
repo.root_group.xarray_tutorial_datasets.air_temperature.air.chunks
/Users/rabernat/mambaforge/envs/arraylake-local/lib/python3.11/site-packages/arraylake/asyn.py:135: UserWarning: You are not on a branch tip, so you can't commit changes.
result[0] = await coro
(730, 7, 14)
repo.checkout("main")
6744c9f10e86f720cdcd34d0
Open Data with Xarray
We can now open our dataset with Xarray.
Arraylake comes with a special to_xarray method to make it very easy to open a particular group as an Xarray dataset.
ds_al = repo.to_xarray("xarray_tutorial_datasets/air_temperature")
print(ds_al)
<xarray.Dataset> Size: 31MB
Dimensions: (time: 2920, lat: 25, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float64 31MB ...
Attributes:
Conventions: COARDS
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...
title: 4x daily NMC reanalysis (1948)
The to_xarray method implements a "fast path" making the dataset quicker to load.
However, it is also possible to use Xarray's own open_dataset or open_zarr functions to load the data. These ultimately produce the same dataset, but can be slower.
ds_al = xr.open_dataset(
repo.store,
group="xarray_tutorial_datasets/air_temperature",
engine="zarr",
zarr_version=3
)
print(ds_al)
<xarray.Dataset> Size: 31MB
Dimensions: (time: 2920, lat: 25, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float64 31MB ...
Attributes:
Conventions: COARDS
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...
title: 4x daily NMC reanalysis (1948)
Modifying Existing Data
As described in detail in the Xarray docs, Xarray supports three different ways of modifying existing Zarr-based datasets. All of these are compatible with Arraylake. We review these three methods below.
Add or Overwrite Entire Variables
To add a new variable, or overwrite an existing variable, in a Zarr-based dataset, use mode='a'.
We'll illustrate this by taking our existing open dataset, renaming the main data variable from air to air_temp and then writing it to the same location in Arraylake.
ds_rename = ds.rename({"air": "air_temp"})
ds_rename.to_zarr(
repo.store,
group="xarray_tutorial_datasets/air_temperature",
mode="a"
)
print(repo.to_xarray("xarray_tutorial_datasets/air_temperature"))
<xarray.Dataset> Size: 31MB
Dimensions: (time: 2920, lat: 25, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 15MB ...
air_temp (time, lat, lon) float32 15MB ...
Attributes:
Conventions: COARDS
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...
title: 4x daily NMC reanalysis (1948)
We can see that we now have two (identical, in this case) data variables with different names. Let's commit this change.
repo.commit("added duplicate data variable with the same name")
65dcae7efb58969e1179efb1
Append to Existing Variables
We often want to extend a dataset along a particular dimension (most commonly time).
To accomplish this, we write new Xarray data to an existing dataset whit the append_dim option set.
To illustrate this, let's split our example dataset into two different parts.
ds_part_1 = ds.isel(time=slice(0, 2000))
ds_part_2 = ds.isel(time=slice(2000, None))
ds_part_1.sizes, ds_part_2.sizes
(Frozen({'lat': 25, 'time': 2000, 'lon': 53}),
Frozen({'lat': 25, 'time': 920, 'lon': 53}))
Now we'll write part 1 to a new dataset:
ds_part_1.to_zarr(
repo.store,
group="xarray_tutorial_datasets/air_temperature_for_appending",
encoding={"air": {"chunks": (200, 25, 53)}}
)
repo.commit("wrote part 1")
65dcae8a477f9574a637100b
Now let's append part 2:
ds_part_2.to_zarr(
repo.store,
group="xarray_tutorial_datasets/air_temperature_for_appending",
append_dim="time"
)
repo.commit("appended part 2")
65dcae99477f9574a637100c
Let's check out the state of the dataset:
ds_appended = repo.to_xarray("xarray_tutorial_datasets/air_temperature_for_appending")
print(ds_appended)
<xarray.Dataset> Size: 15MB
Dimensions: (time: 2920, lat: 25, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 15MB ...
Attributes:
Conventions: COARDS
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...
title: 4x daily NMC reanalysis (1948)
Great! We've reconstructed the original dataset by writing it incrementally in two separate commits.
Let's just double check that the final dataset is exactly as it should be.
ds.identical(ds_appended)
True
Write Limited Regions of Existing Datsets
This final option is useful when you have a large dataset, whose size is known in advance, which you want to populate by writing many smaller pieces at a time.
To use this option, first we need to create a new dataset to serve as a "template" or "schema" for the full dataset. For this example, we'll manually create a global lat-lon grid, and then "insert" our example dataset into it.
Here we use the trick of creating a dummy Dask array to stand in for the data we will be writing later. We'll use the same resolution as our original dataset (2.5 degrees).
import dask.array as da
import numpy as np
lat = np.arange(-90, 90, 2.5)
lon = np.arange(0, 360, 2.5)
ny, nx = len(lat), len(lon)
nt = ds.sizes['time']
ds_template = xr.Dataset(
data_vars={
"air": (
('time', 'lat', 'lon'),
da.zeros((nt, ny, nx), chunks=(200, ny, nx), dtype=ds.air.dtype)
)
},
coords={
'time': ds.time,
'lat': lat,
'lon': lon
}
)
print(ds_template)
<xarray.Dataset> Size: 121MB
Dimensions: (time: 2920, lat: 72, lon: 144)
Coordinates:
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
* lat (lat) float64 576B -90.0 -87.5 -85.0 -82.5 ... 80.0 82.5 85.0 87.5
* lon (lon) float64 1kB 0.0 2.5 5.0 7.5 10.0 ... 350.0 352.5 355.0 357.5
Data variables:
air (time, lat, lon) float32 121MB dask.array<chunksize=(200, 72, 144), meta=np.ndarray>
To initialize the target dataset in arraylake, we store it with the compute=False option.
This means that only the coordinates will be written.
ds_template.to_zarr(repo.store, group="global_air_temp", compute=False)
repo.commit("Wrote template dataset")
65dcaeaaf5b1f8db1b6f1bea
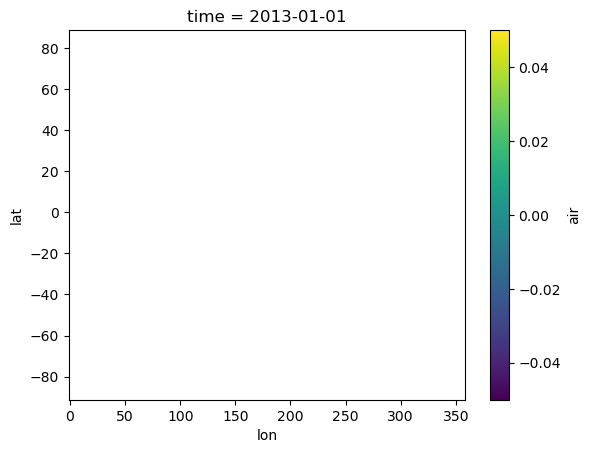
As we can see, this dataset is currently empty.
repo.to_xarray("global_air_temp").air.isel(time=0).plot()
<matplotlib.collections.QuadMesh at 0x285b365d0>
Now we will write our dataset to it using the region="auto" option.
Xarray will automatically determine the correct alignment with the target dataset and insert it in the right place.
(For more manual control, explicit slices can also be passed; see Xarray API documentation for more detail.)
One minor trick is required here. Our original dataset has data ordered with decreasing lats. We need to reverse these to match the increasing order in our template dataset.
ds_reversed = ds.isel(lat=slice(None, None, -1))
ds_reversed.to_zarr(repo.store, group="global_air_temp", region="auto")
repo.commit("wrote region")
65dcaecaf5b1f8db1b6f1beb
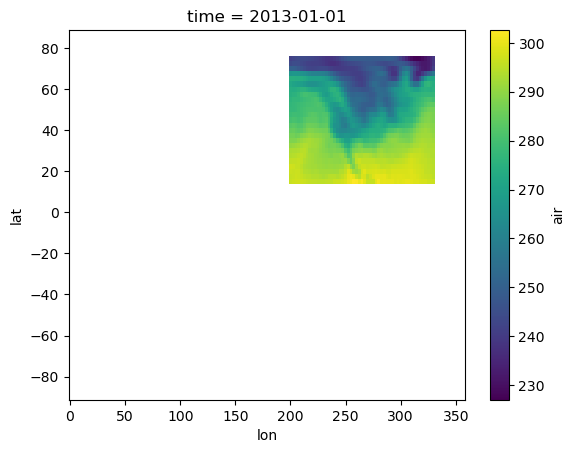
ds_global = repo.to_xarray("global_air_temp")
ds_global.air.isel(time=0).plot()
<matplotlib.collections.QuadMesh at 0x285cb1b10>
To round out this example, let's make a nice map.
import cartopy.crs as ccrs
from matplotlib import pyplot as plt
fig, ax = plt.subplots(subplot_kw={"projection": ccrs.Robinson()})
ds_global.air[0].plot(transform=ccrs.PlateCarree())
ax.coastlines()
<cartopy.mpl.feature_artist.FeatureArtist at 0x285acfcd0>
/Users/rabernat/mambaforge/envs/arraylake-local/lib/python3.11/site-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/110m_physical/ne_110m_coastline.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
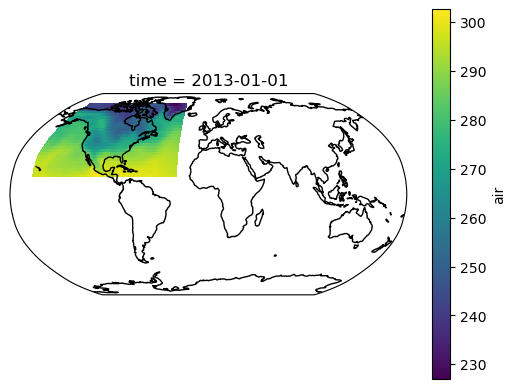
This method can be scaled out to populate very large spatial domains one piece at a time.
When writing to regions from a single process, you can write any region you want. However, when writing to regions concurrently, you may need to consider the alignment of each worker's write region with the underlying chunks. See scaling for more information on concurrent writes to Arraylake.
Combining Multiple Datasets
A popular feature in Xarray is the open_mfdataset function, which automatically reads and combines many files into a single Dataset. open_mfdataset doesn't work with Arraylake. However, it's easy to achieve similar results.
Arraylake users have two good options:
- Just use one single Dataset from the beginning. Arraylake can efficiently handle datasets of any size (up to Petabytes). Because it's easy to extend data in Arraylake, you don't necessarily need to split your data into many small individual Datasets. This way of thinking about data is a relic of old-fashioned file-based workflows.
- Open your datasets individually and then manually combine them. This is exactly what
open_mfdatasetdoes under the hood anyway.
Here we illustrate option 2 using the same two-part dataset we created above. For more information, see the Xarray docs on combining data.
First we write each of these to a separate Arraylake dataset.
ds_part_1.to_zarr(repo.store, group="air_temp/part_1")
ds_part_2.to_zarr(repo.store, group="air_temp/part_2")
repo.commit("wrote two datasets")
65dcaef55465a099fc27bbca
Now we will open each back up and combine them.
dsets = [repo.to_xarray("air_temp/part_1"), repo.to_xarray("air_temp/part_2")]
ds_combined = xr.combine_by_coords(dsets)
print(ds_combined)
<xarray.Dataset> Size: 15MB
Dimensions: (time: 2920, lat: 25, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 15MB 241.2 242.5 243.5 ... 296.2 295.7
Attributes:
Conventions: COARDS
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...
title: 4x daily NMC reanalysis (1948)
As we can see, the two datasets have been combined back into one single dataset.
To wrap up this tutorial, we will clean up by deleting this repo we just created.
client.delete_repo("earthmover/xarray-tutorial-datasets", imsure=True, imreallysure=True)